Model Specification and Building¶
The specification and building of multi-cellular system models in Infobiotics is modular allowing parsimonious and incremental design. In this chapter, the Infobiotics modelling language is introduced using a running example consisting of the synthetic bacterial colony designed by Ron Weiss’ group in [Basu2005]. This model implements the propagation of a wave of gene expression in a bacterial colony.
The Infobiotics modelling language provides a multi-compartmental, stochastic and rule-based specification framework. A model of a multi-cellular system in Infobiotics is developed as a Lattice Population P-system (LPP-system) which consists of the specification of three main components that can be defined in a modular manner:
- First, the different cell types in the multi-cellular system need to be specified including their molecular species, compartmentalised structure and molecular interactions.
- Second, the geometric distribution of the cells in the multi-cellular system has to be captured using a finite point lattice, a regularly distributed collection of spatial points.
- Finally, the specific localisation of the different cells over the lattice points must be described in order to obtain the final spatial distribution in the multi-cellular system.
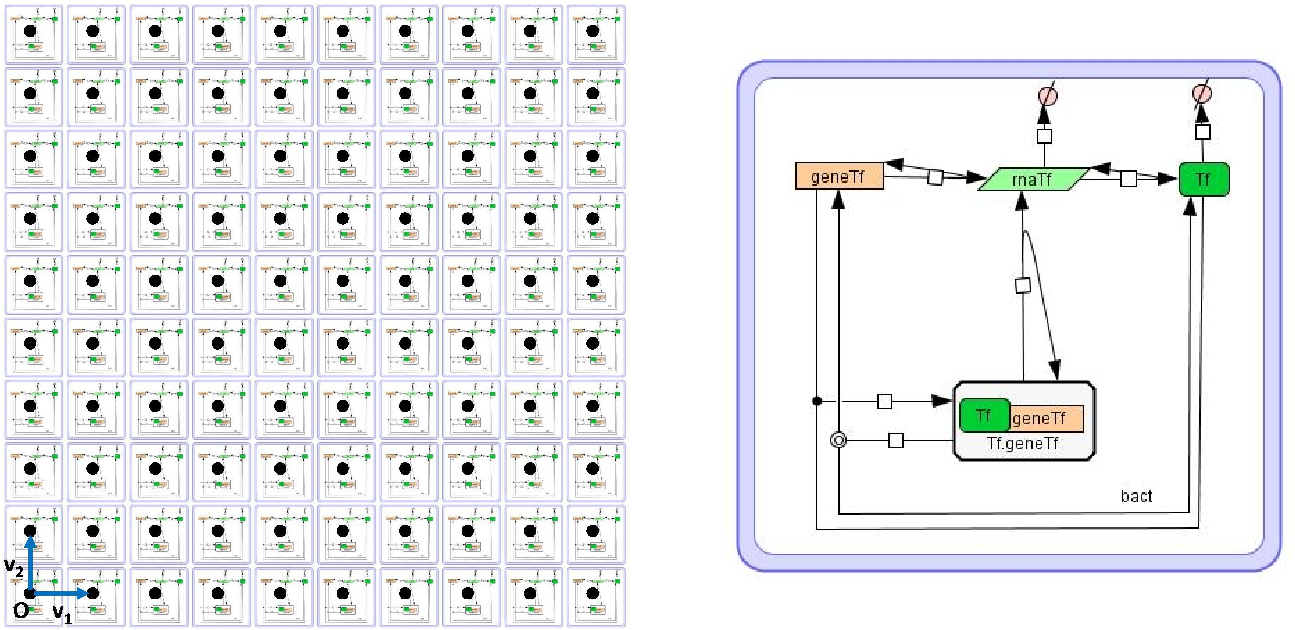
Example of an LPP-system consisting of the distribution of the model of a single cell containing a negative autoregulated gene (right) over a regular rectangular lattice (left).
The specification of the different parts of a multicelluar system using the Infobiotics modelling language is specified in the following sections:
- Specification of Cell Types
- Specification of Geometric Distribution
- Specification of Multi-cellular Systems
Credits:
The Infobiotics modelling language was developed by Francisco J. Romero-Campero with contributions from Jamie Twycross, Jonathan Blakes and Hongqing Cao. It is being used on Systems and Synthetic Biology research projects in the University of Nottingham, U.K.