Model Specification and Building¶
The Infobiotics modelling language is based on Stochastic P-systems, SP-system for short. It allows you to specify your models in a parsimonious and incremental way. Typically, multi-cellular models are specified in our modelling language using libraries of re-usable modules, models of individual cell types and geometric distributions of clones of theses cell types. We will illustrate this using our running example based on gene autoregulation.
Libraries of reusable modules¶
Libraries of modules must be specified using text files with the extension plb. You can open the file containing the library in our running example, basicLibrary.plb, by selecting File -> Open text file from the upper menu bar and navigating to its specific location.
Our libraries are specified using the key workds libraryOfModules ... endLibraryOfModules. Each library is identified with a name, basicLibrary in our running example and consists of a collection of modules of molecular interactions. In our case we have three different modules, UnReg, PosReg and NegReg describing constitutive gene expression, gene positive regulation and gene negative regulation respectively. Comments can be included using text enclosed between # symbols. Below you have the library used in the autoregulation example:
# Author: Francisco J. Romero-Campero #
# Date: May 2010 #
# Description: A library containing basic gene regulatory mechanisms #
libraryOfModules basicLibrary
# A module representing the unregulated expression of a gene X #
UnReg({X},{c_1, c_2, c_3, c_4},{l}) =
{
rules:
# Transcription of geneX #
r1: [ geneX ]_l -c_1-> [ geneX + rnaX ]_l
# Degradation of the RNA #
r2: [ rnaX ]_l -c_2-> [ ]_l
# Translation of the RNA #
r3: [ rnaX ]_l -c_3-> [ rnaX + proteinX ]_l
# Degradation of the protein #
r4: [ proteinX ]_l -c_4-> [ ]_l
}
# A module representing the positive regulation of a protein X over a gene Y #
PosReg({X,Y},{c_1,c_2,c_3,c_4,c_5,c_6},{l}) =
{
rules:
# Binding and debinding of the transcription factor proteinX to geneY #
r1: [ proteinX + geneY ]_l -c_1-> [ proteinX_geneY ]_l
r2: [ proteinX_geneY ]_l -c_2-> [ proteinX + geneY ]_l
# Transcription of geneY when proteinX is bound to its promoter #
r3: [ proteinX_geneY ]_l -c_3-> [ proteinX_geneY + rnaY ]_l
r4: [ rnaY ]_l -c_4-> [ ]_l
r5: [ rnaY ]_l -c_5-> [ rnaY + proteinY ]_l
r6: [ proteinY ]_l -c_6-> [ ]_l
}
# A module representing the negative regulation of a protein X over a gene Y #
NegReg({X,Y},{c_1,c_2},{l}) =
{
rules:
# Binding and debinding of the transcription factor proteinX to gene Y #
r1: [ proteinX + geneY ]_l -c_1-> [ proteinX_geneY ]_l
r2: [ proteinX_geneY ]_l -c_2-> [ proteinX + geneY ]_l
}
endLibraryOfModules
A module is identified with a name and is associated three sets of variables, i.e. UnReg({X},{c_1, c_2, c_3, c_4},{l}). The first set of variables can be instantiated with names of specific molecular species. The second set of variables can be instantiated with numerical values capturing the rates of the molecular interactions represented in the module. Finally, the third set of variables can be instantiated with the names of the compartments involved in the molecular interactions.
A module encapsulates a set of molecular interactions represented by rules (or other modules) that may use some of the module variables. For example, rules r1 and r2 from the NegReg module describe the binding/debiding of proteinX to/from geneY. These processes take place at rates c_1 and c_2 inside compartment l represented using square brackets. Note that X, Y, c_1, c_2 and l are module variables that can be instantiated with specific values:
r1: [ proteinX + geneY ]_l -c_1-> [ proteinX_geneY ]_l
r2: [ proteinX_geneY ]_l -c_2-> [ proteinX + geneY ]_l
Other attributes can be associated with our modules such as types and specific DNA sequences see this example or the documentation for more details.
Cell types¶
The Infobiotics modelling language supports the specification of cell types in two different formats. You can specify your cell types using SBML, this example. Alternatively, if you want to use modules of molecular interactions from different libraries defined previously you need to specify your cell type using our modelling language. In this last case, you must use a text file with the extension sps, from SP-system.
Our autoregulation example uses three different cell types carrying the same gene under three different regulatory mechanisms, unregulated expression (UnReg.sps), positive autoregulation (PAR.sps) and negative autoregulation (NAR.sps). Open the model of the last cell type by selecting File -> Open text file from the upper menu bar and choosing the file NAR.sps:
# Author: Francisco J. Romero-Campero #
# Date: July 2010 #
# Description: A model of a cell type carrying a gene that regulates itself negatively #
SPsystem negativeAutoregulation
# Molecular species in the system #
alphabet
gene1
protein1
protein1_gene1
rna1
signal1
endAlphabet
# The system consists of two compartments called media and bacterium. The bacterium #
# is embedded in the media #
compartments
media
bacterium inside media
endCompartments
# The initial number of molecules present in the system #
initialMultisets
initialMultiset bacterium
gene1 1
endInitialMultiset
endInitialMultisets
# The rules describing the molecular interactions in the different compartments #
# of the system #
ruleSets
# Molecular interactions involving the compartment bacterium #
ruleSet bacterium
# Unregulated expression of gene 1 #
UnReg({1},{3,0.07,3,0.01},{bacterium}) from basicLibrary.plb
# Negative regulation of gene 1 by the protein product of gene 1 #
NegReg({1,1},{1,0.8},{bacterium}) from basicLibrary.plb
# Protein1 is an enzyme that synthesizes signal 1 #
r1: [ protein1 ]_bacterium -c1-> [ protein1 + signal1 ]_bacterium c1 = 0.001
# Signal1 diffuses freely outside bacteria #
r2: [ signal1 ]_bacterium -c2-> signal1 [ ]_bacterium c2 = 0.001
# Singal1 can be degraded inside bacteria #
r3: [ signal1 ]_bacterium -c3-> [ ]_bacterium c3 = 0.0001
endRuleSet
# Molecular interactions involving the compartment media #
ruleSet media
# Signal1 diffuses freely inside bacteria #
r1: signal1 [ ]_bacterium -c1-> [ signal1 ]_bacterium c1 = 0.001
# Signal1 can be degraded in the media #
r2: [ signal1 ]_bacterium -c2-> [ ]_bacterium c2 = 0.0001
# Signal1 diffuses freely to neighbouring media #
r3: [ signal1 ]_bacterium =(1,0)=[ ] -c3-> [ ]_bacterium =(1,0)=[ signal1 ] c3 = 0.00025
r4: [ signal1 ]_bacterium =(-1,0)=[ ] -c3-> [ ]_bacterium =(-1,0)=[ signal1 ] c3 = 0.00025
r5: [ signal1 ]_bacterium =(0,1)=[ ] -c3-> [ ]_bacterium =(0,1)=[ signal1 ] c3 = 0.00025
r6: [ signal1 ]_bacterium =(0,-1)=[ ] -c3-> [ ]_bacterium =(0,-1)=[ signal1 ] c3 = 0.00025
endRuleSet
endRuleSets
endSPsystem
- A cell type is specified using the key words SPsystem ... endSPsystem. Each cell type is identified with a name, negativeAutoregulation in our example above, and specifies the following components of a single-cell model:
- The molecular species in the cell type are specified as a list of names (gene1, protein1, etc) enclosed between the key words alphabet ... endAlphabet.
- The compartments of the cell type are listed using the key words compartments ... endCompartments. Each compartment is specified using its name. If a compartment is embedded in another one the key word inside is used. For instance, the cell type above consists of two compartments, media and bacterium. The compartment bacterium is contained in the media which is specified as bacterium inside media.
- The initial number of molecules is specified with the key words initialMultisets ... endInitialMultisets. Each comparmtent is specifically associated with its initial number of molecules using the key words initialMultiset ... endInitialMultiset and its name. The number of molecules is specified as a list of molecular species names followed by a positive integer number. In our example, only a single copy of gene1 is initially present in the compartment bacterium.
- The molecular interactions taking place inside or between compartments are enumerated within the key words ruleSets ... endRuleSets. The molecular interactions associated with a compartment with name CompName are specified as a list of rules and instantiated modules enclosed between the key words ruleSet ... endRuleSet and identified with CompName. The library file where the modules are defined must be specified after the module instantiation using the key word from. In our example, the interactions involving the compartment bacterium are specified using two instantiated modules defined in the basicLibrary.plb file (UnReg({1},{3,0.07,3,0.01},{bacterium}) and NegReg({1,1},{1,0.8},{bacterium})) and three rules describing the synthesis of singal1 by protein1 (rule r1), the diffusion of singal1 outside the bacterium (rule r2) and the degradation of signal1 (rule r3). The values of the stochastic constants associated with these rules are also stated at the end of the rule specifications . Note that the outtermost compartment of a cell type can diffuse molecules to neighbouring cells using rules of the same form as rules r3, r4, r5 and r6 in the media compartment. A vector v is associated with this type of rule. The application of a rule of this form in a cell located at position p moves the corresponding molecules to the cell located at position p+v.
Geometric distribution¶
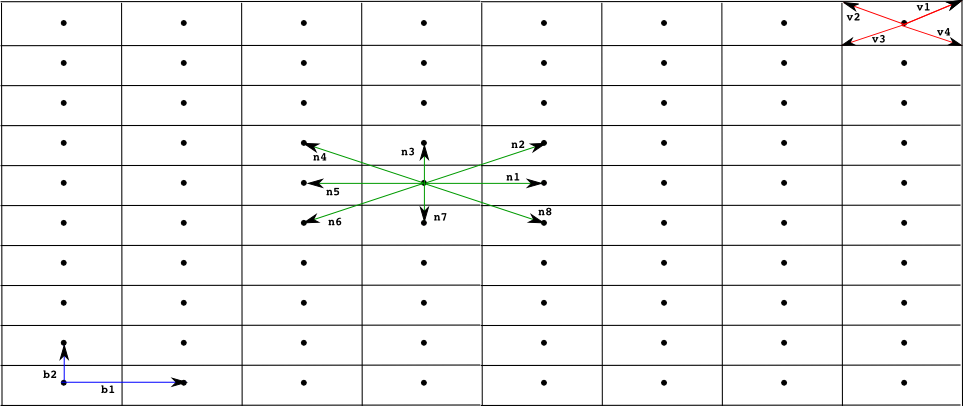
The Infobiotics modelling language allows you to specify the spatial distribution of cells in multi-cellular systems. The characteristic geometry of such systems is captured using finite point lattices (a grid of regularly distributed spatial points) that must be specified in files with the extension lat. Our running example uses a rectangular lattice similar to the one shown below.
Open the rectangular lattice used in our example by selecting File -> Open text file from the upper menu bar and choosing the file rectangular.lat:
# Author: Francisco J. Romero-Campero #
# Date: July 2010 #
# Description: A rectangular lattice of size 40x10 #
lattice rectangularLattice
# Dimension of the lattice and lower/upper bounds #
dimension 2
xmin 0
xmax 39
ymin 0
ymax 9
# Parameters used in the definition of the rest of components defining the lattice #
parameters
parameter b1 value = 2
parameter b2 value = 1
endParameters
# Basis vector determining the points in the lattice #
# in this case we have a rectangular lattice #
basis
(b1,0)
(0,b2)
endBasis
# Vertices used to determine the shape of the outmost membrane #
# of the SP systems located on each point of the lattice #
vertices
(b1/2,b2/2)
(-b1/2,b2/2)
(-b1/2,-b2/2)
(b1/2,-b2/2)
endVertices
# Vectors pointing at the neighbours of each point of the lattice #
neighbours
(1,0) (1,1) (0,1) (-1,1)
(-1,0) (-1,-1) (0,-1) (1,-1)
endNeighbours
endLattice
- A lattice is specified using the key words lattice ... endLattice and identified with a name, rectangularLattice in our example. The specification of a lattice consists of the following components:
- Currently, our modelling language supports only one and two dimensional lattices. This must be specified after the key word dimension.
- The lattice size is determined by the lower and upper bounds specified using the key words xmin, xmax, ymin and ymax followed by positive integer values.
- A set of parameters can be used in the lattice specification. These are introduced using the key words parameters ... endParameters. Each parameter is identified with a name and is given a value.
- The points of a lattice are determined by a set of basis vectors that are listed within the key words basis ... endBasis. The lattice points are then obtained as all the possible linear combinations of the basis vectors with integer coefficients within the given bounds.
- A regular polygon, a rectangle in our example, is associated with each lattice point to specify the shape of the cell located in that position. The vertices of the polygon are computed using a list of vectors introduced using the key words vertices ... endVertices.
- A neighbourhood is associated with each lattice point. This is specified as a list of vectors within the key words neighbours ... endNeighbours.
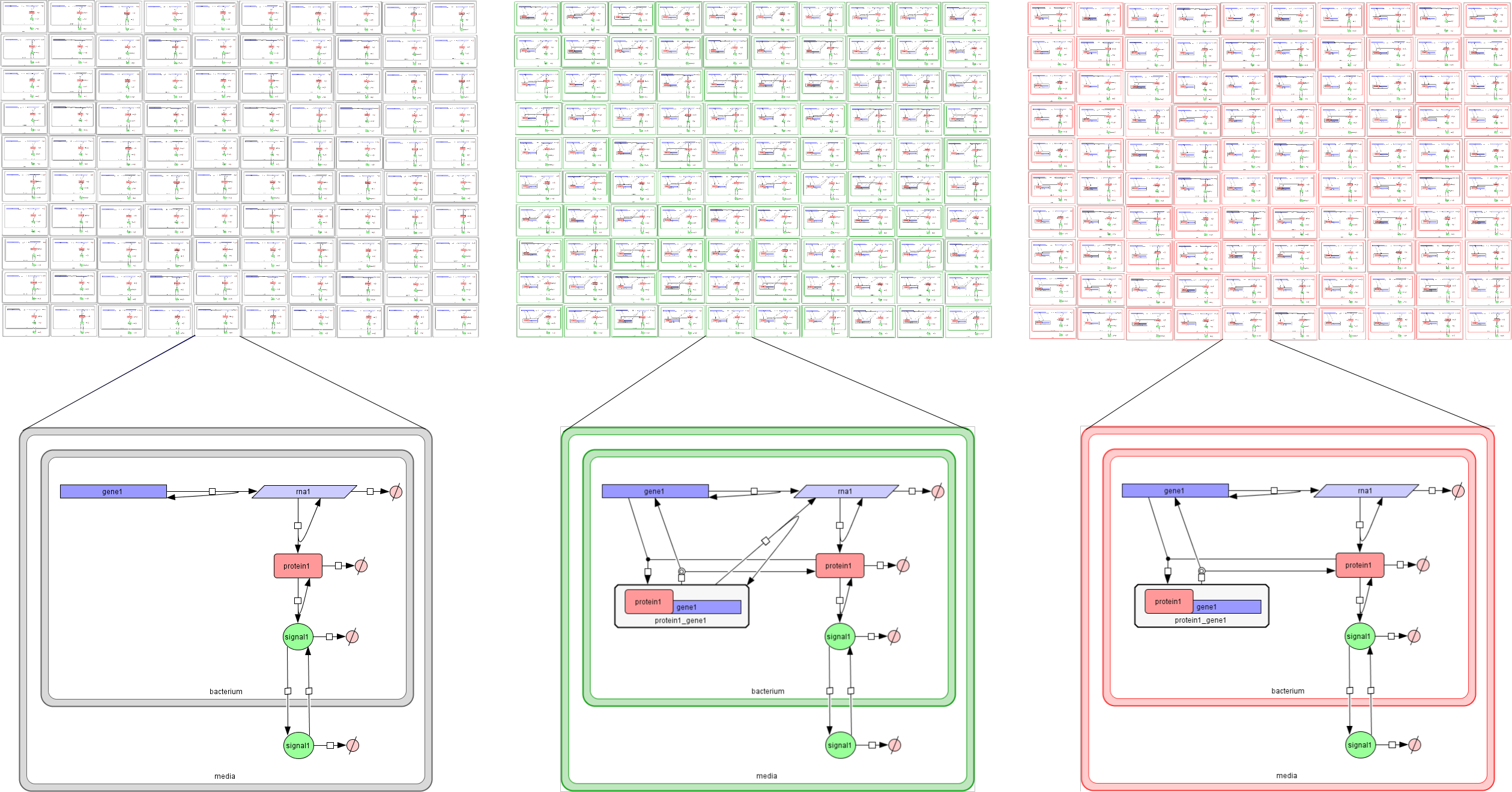
A model of a multi-cellular system in our modelling language consists of a Lattice Population P-system, LPP-system for short, a distribution of many clones of the different cell types represented as SP-systems over lattice points. These models must be specified in text files with the extension lpp. Open the model of our multi-cellular system consisting of three bacterial colonies by selecting File -> Open text file from the upper menu bar and choosing the file bacterialColonies.lpp:
# Author: Francisco J. Romero-Campero #
# Date: July 2010 #
# Description: A multicelluar system consisting of three bacterial colonies #
# combining bacteria carrying the same gene under three different #
# regulatory mechanisms. Namely, unregulated expression, positive #
# autoregulation and negative autoregulation #
LPPsystem threeColonies
# Cell types specified as individual SP systems #
SPsystems
SPsystem UnReg from UnReg.sps
SPsystem PAR from PAR.sps
SPsystem NAR from NAR.sps
endSPsystems
# The geometry of the system is represented using a regular finite point lattice #
lattice rectangular from rectangular.lat
# Special distribution of the cells over the lattice points#
spatialDistribution
# Bacteria carrying gene1 expressed constitutively are place on the left #
positions for UnReg
parameters
parameter i = 0:1:9
parameter j = 0:1:9
endParameters
coordinates
x = i
y = j
endCoordinates
endPositions
# Bacteria carrying gene1 regulating itself positively are place at the center #
positions for PAR
parameters
parameter i = 15:1:24
parameter j = 0:1:9
endParameters
coordinates
x = i
y = j
endCoordinates
endPositions
# Bacteria carrying gene1 regulating itself negatively are place on the right #
positions for NAR
parameters
parameter i = 30:1:39
parameter j = 0:1:9
endParameters
coordinates
x = i
y = j
endCoordinates
endPositions
endSpatialDistribution
endLPPsystem
A multi-cellular model is specified using the key words LPPsystem ... endLPPsystem and is identified with a name, threeColonies, in our example. The specification of a multi-cellular model consists of the following components:
- A list of cell types defined as SP-systems introduced within the key words SPsystems ... endSPsystems. Each cell type is specified using the key word SPsystem followed by the name given to the cell type, the key word from and the file where the single cell model is specified. Recall that single-cell models can be specified in sps format as described above or in SBML format. Our autoregulation example uses three different cell types UnReg, PAR and NAR introduced in the files UnReg.sps, PAR.sps and NAR.sps respectively.
- The declaration of the geometry of the system uses the key word lattice followed by a name, the key word from and the file where the corresponding finite point lattice is specified. Our example uses a rectangular lattice described in the rectangular.lat file.
- The spatial distribution of cellular clones over the lattice points. This is specified using the key words spatialDistribution ... endSpatialDistribution. A list of positions associated to each cell type is introduced using the key words positions for ... endPositions specifying the name of the corresponding SP-system. These positions can use some parameters with specific ranges described using the key word parameter followed by the parameter name, the equal symbol and the lower bound, step and upper bound separated by : symbols. Finally the coordinates, x and y, are specified as mathematical expressions that may use the previously introduced parameters.
Below you have a figure representing the autoregulation model with its specific cell types and spatial distribution.