Automatic Discovery of Pulse Generators¶
Introduction¶
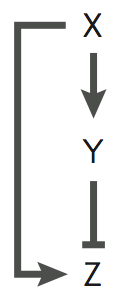
The feed-forward loop (FFL) is a well studied network motif is systems biology. This model implements a pulse generator based on an incoherent type-1 FFL:
For this common FFL motif the transcription activator X activates gene Z directly, and also activates the repressor Y of the gene Z, generating a pulse on the expression of Z. The complete model can be downloaded from this link.
Library of Modules¶
The library of modules defines all modules that can be part of candidate models. This is essentially the list of buildings blocks from where the model will be constructed:
libraryOfModules FFL_libray
# A module representing the unregulated expression of a gene X #
UnReg({X},{c_1 0:0.1:10 linear, c_2 0:0.01:2 linear, c_3 0:0.01:2 linear, c_4 0:0.01:2 linear},{l}) =
{
rules:
# Transcription of geneX #
r1: [ geneX ]_l -c_1-> [ geneX + rnaX ]_l
# Degradation of the RNA #
r2: [ rnaX ]_l -c_2-> [ ]_l
# Translation of the RNA #
r3: [ rnaX ]_l -c_3-> [ rnaX + proteinX ]_l
# Degradation of the protein #
r4: [ proteinX ]_l -c_4-> [ ]_l
}
# A module representing the positive regulation of a protein X #
# over a gene Y #
PosReg({X,Y},{c_1 0:0.1:10 linear,c_2 -3:1:3 logarithmic,c_3 0:0.1:10 linear, c_4 0:0.01:2 linear, c_5 0:0.01:2 linear, c_6 0:0.01:2 linear},{l}) =
{
rules:
# Binding and debinding of the transcription factor proteinX to geneY #
r1: [ proteinX + geneY ]_l -c_1-> [ proteinX_geneY ]_l
r2: [ proteinX_geneY ]_l -c_2-> [ proteinX + geneY ]_l
# Transcription of geneY when proteinX is bound to its promoter #
r3: [ proteinX_geneY ]_l -c_3-> [ proteinX_geneY + rnaY ]_l
# Degradation of the RNA #
r4: [ rnaY ]_l -c_4-> [ ]_l
# Translation of the RNA #
r5: [ rnaY ]_l -c_5-> [ rnaY + proteinY ]_l
# Degradation of the protein #
r6: [ proteinY ]_l -c_6-> [ ]_l
}
# A module representing the negative regulation of a protein X #
# over a gene Y #
NegReg({X,Y},{c_1 0:0.1:10 linear, c_2 -3:1:3 logarithmic},{l}) =
{
rules:
# Binding and debinding of the transcription factor proteinX to gene Y #
r1: [ proteinX + geneY ]_l -c_1-> [ proteinX_geneY ]_l
r2: [ proteinX_geneY ]_l -c_2-> [ proteinX + geneY ]_l
}
endLibraryOfModules
Target Model¶
The target model is composed by the following modules:
UnReg(X=gene1)
PosReg(X=gene1,Y=gene2)
PosReg(X=gene1,Y=gene3)
NegReg(X=gene2,Y=gene3)
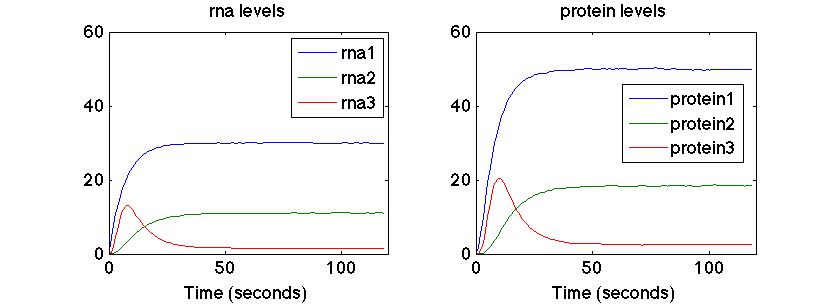
The model is optimised to output the target behaviour given by the time series for the rna and protein levels. The fitness is calculated based on the RMSE between the model output and the target data. As you can see below the expression of gene 3 generates a pulse.
Optimisation¶
The optimisation procedure can take some hours depending on the dimension of the search space. This include number of modules, possible instantiations for each module, and number of associated parameters (along with their ranges and precision). To speedup this process you can relax some of the optimisation parameters (population size, number of generations, number of simulation runs, proportion of models for parameter optimisation) at the cost of less accurate final models.