Pulse Generator¶
Introduction¶
The pulse generator example consists of the synthetic bacterial colony designed by Ron Weiss’ group in [Basu2004,Basu2005]. This model implements the propagation of a wav gene expression in a bacterial colony. The complete model and experimental results of can be downloaded from this link.
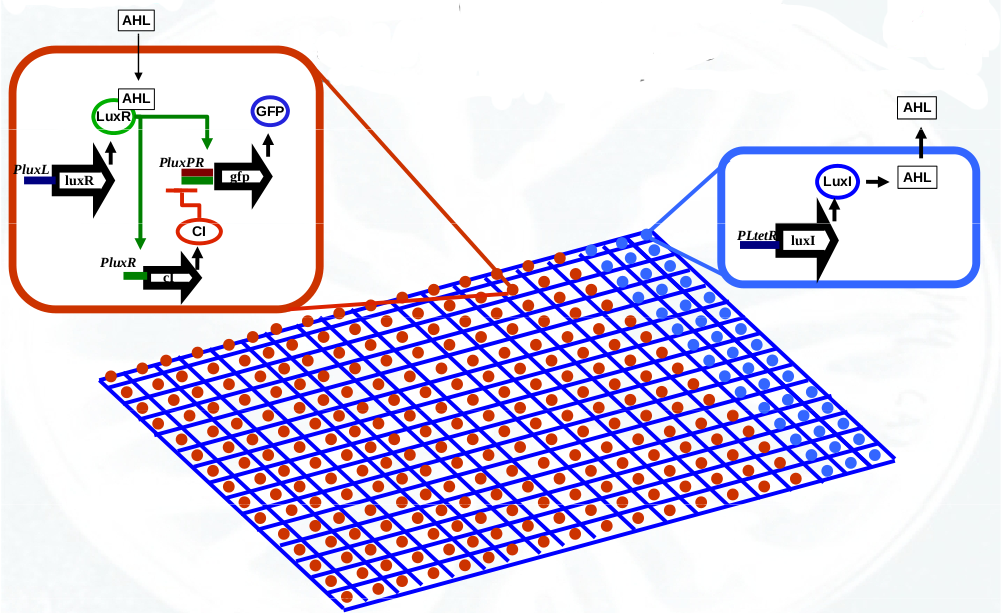
The pulse generator consists of two different bacterial strains, sender cells and pulsing cells:
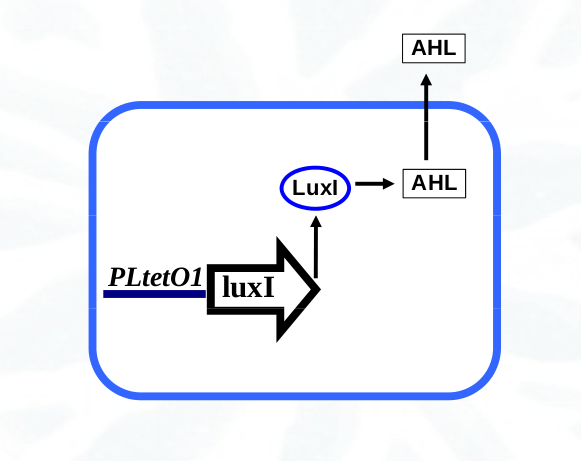
- Sender cells contain the gene luxI from Vibrio fischeri. This gene codifies the enzyme LuxI responsible for the synthesis of the molecular signal 3OC6-HSL (AHL). The luxI gene is expressed constitutively under the regulation of the promoter PLtetO1 from the tetracycline resistance transposon.
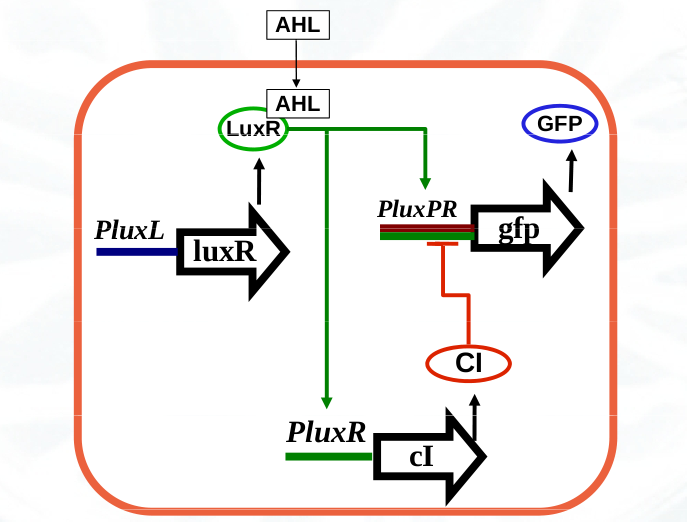
- Pulsing cells contain the luxR gene from Vibrio fischeri that codifies the 3OC6-HSL receptor protein LuxR. This gene is under the constitutive expression of the promoter PluxL from Vibrio fischeri. It also contains the gene cI from lambda phage codifying the repressor CI under the regulation of the promoter PluxR that is activated upon binding of the transcription factor LuxR_3OC6_2. Finally, this bacterial strain carries the gene GFP that codifies the green fluorescent protein under the regulation of the synthetic promoter PluxPR combining the Plux promoter (activated by the transcription factor LuxR_3OC6_2) and the PR promoter from lambda phage (repressed by the transcription factor CI).
The bacterial strains above are distributed in a specific spatial distribution. Sender cells are located at one end of the bacterial colony and the rest of the system is filled with pulsing cells as the figure below shows:
The Model¶
Our model of the Pulse Generator uses a module library describing the regulation of the different promoters used in the two bacterial strains. This library is presented below:
# Author: Francisco J. Romero-Campero #
# Date: February 2010 #
# Description: This module library describes the regulation of the gene promoters used in the #
# pulse generator circuit developed by Ron Weiss group #
libraryOfModules promoterLibrary
# This module represents the constitutive expression of a gene X under the regulation #
# of the promoter PLtetO1 from the tetracycline resistance transposon #
PLtetO1({X},{c_1},{l}) =
{
# This module represents a promoter #
type: promoter
# DNA sequence corresponding to biobrick BBa_R0040 from the Registry of Standard Biological Parts #
sequence: TCCCTATCAGTGATAGAGATTGACATCCCTATCAGTGATAGAGATACTGAGCAC
rules:
r1: [ PLtetO1_geneX ]_l -c_1-> [ PLtetO1_geneX + rnaX_RNAP ]_l
}
# This module represents the constitutive expression of a gene X under the regulation #
# of the promoter PluxL from Vibrio fischeri #
PluxL({X},{c_1},{l}) =
{
# This module represents a promoter #
type: promoter
# DNA sequence corresponding to biobrick BBa_R0063 from the Registry of Standard Biological Parts #
sequence: ACCTGTACGATCCTACAGGTGCTTATGTTAAGTAATTGTATTCCCAGCGATACAATAGTGTGACAAAAATCCAAT
TTATTAGAATCAAATGTCAATCCATTACCGTTTTAATGATATATAACACGCAAAACTTGCGACAAACAATAGGTA
rules:
r1: [ PluxL_geneX ]_l -c_1-> [ PluxL_geneX + rnaX_RNAP ]_l
}
# This module represents the positively regulated expression of a gene X under the control #
# of the promoter PluxR which is activated by LuxR_2 #
PluxR({X},{c_1, c_2, c_3},{l}) =
{
# This module represents a promoter #
type: promoter
# DNA sequence corresponding to biobrick BBa_R0062 from the Registry of Standard Biological Parts #
sequence: ACCTGTAGGATCGTACAGGTTTACGCAAGAAAATGGTTTGTTATAGTCGAATAAA
rules:
r1: [ LuxR2 + PluxR_geneX ]_l -c_1-> [ PluxR_LuxR2_geneX ]_l
r2: [ PluxR_LuxR2_geneX ]_l -c_2-> [ LuxR2 + PluxR_geneX ]_l
r3: [ PluxR_LuxR2_geneX ]_l -c_3-> [ PluxR_LuxR2_geneX + rnaX_RNAP ]_l
}
# This module represents the positivele/negatively regulated expression of a gene X under the control #
# of the synthetic promoter PluxPR that combines the activation by LuxR2 with the repression by CI2 #
PluxPR({X},{c_1,c_2,c_3,c_4,c_5,c_6,c_7,c_8,c_9},{l}) =
{
# This module represents a promoter #
type: promoter
# DNA sequence corresponding to biobrick BBa_I1051 from the Registry of Standard Biological Parts #
sequence: ACCTGTAGGATCGTACAGGTTTACGCAAGAAAATGGTTTGTTATAGTCGAATACCTCTGGCGGTGATA
rules:
r1: [ LuxR2 + PluxPR_geneX ]_l -c_1-> [ PluxPR_LuxR2_geneX ]_l
r2: [ PluxPR_LuxR2_geneX ]_l -c_2-> [ LuxR2 + PluxPR_geneX ]_l
r3: [ LuxR2 + PluxPR_CI2_geneX ]_l -c_3-> [ PluxPR_LuxR2_CI2_geneX ]_l
r4: [ PluxPR_LuxR2_CI2_geneX ]_l -c_4-> [ LuxR2 + PluxPR_CI2_geneX ]_l
r5: [ CI2 + PluxPR_geneX ]_l -c_5-> [ PluxPR_CI2_geneX ]_l
r6: [ PluxPR_CI2_geneX ]_l -c_6-> [ CI2 + PluxPR_geneX ]_l
r7: [ CI2 + PluxPR_LuxR2_geneX ]_l -c_7-> [ PluxPR_LuxR2_CI2_geneX ]_l
r8: [ PluxPR_LuxR2_CI2_geneX ]_l -c_8-> [ CI2 + PluxPR_LuxR2_geneX ]_l
r9: [ PluxPR_LuxR2_geneX ]_l -c_9-> [ PluxPR_LuxR2_geneX + rnaX_RNAP ]_l
}
endLibraryOfModules
An additional module library describing several post-transcriptional regulatory mechanisms is also used in our model:
# Author: Francisco J. Romero-Campero #
# Date: February 2010 #
# Description: This module library describes some general post-transcriptional regulatory mechanisms #
libraryOfModules postTrasncriptionalRegulation
# This module represents transcription termination, translation, rna and protein degradation #
PostTransc({X},{c_1,c_2,c_3,c_4,c_5},{l}) =
{
rules:
r1: [ rnaX_RNAP ]_l -c_1-> [ rnaX ]_l
r2: [ rnaX ]_l -c_2-> [ rnaX + proteinX_Rib ]_l
r3: [ rnaX ]_l -c_3-> [ ]_l
r4: [ proteinX_Rib ]_l -c_4-> [ proteinX ]_l
r5: [ proteinX ]_l -c_5-> [ ]_l
}
# This module represents the dimerisation of a protein X #
Dim({X,Y},{c_1,c_2},{l}) =
{
rules:
r1: [ proteinX + proteinX ]_l -c_1-> [ Y ]_l
r2: [ Y ]_l -c_2-> [ ]_l
}
# This module represents the dimerisation of a protein X in the presence of a signal S #
DimSig({X,S,Y},{c_1,c_2,c_3,c_4},{l}) =
{
rules:
r1: [ proteinX + signalS ]_l -c_1-> [ proteinX_S ]_l
r2: [ proteinX_S ]_l -c_2-> [ ]_l
r3: [ proteinX_S + proteinX_S ]_l -c_3-> [ Y ]_l
r4: [ Y ]_l -c_4-> [ ]_l
}
# This module represents the free diffusion of a singal X in a rectangular latice #
Diffusion({X},{c_1},{l}) =
{
rules:
r1: [ signalX ]_l =(1,0)=[ ] -c_1-> [ ]_l =(1,0)=[ signalX ]
r2: [ signalX ]_l =(-1,0)=[ ] -c_1-> [ ]_l =(-1,0)=[ signalX ]
r3: [ signalX ]_l =(0,1)=[ ] -c_1-> [ ]_l =(0,1)=[ signalX ]
r4: [ signalX ]_l =(0,-1)=[ ] -c_1-> [ ]_l =(0,-1)=[ signalX ]
}
endLibraryOfModules
The bacterial strain, senderCell, producing the signal 3OC6-HSL (AHL) is modelled using the SP-system model below:
# Author: Francisco J. Romero-Campero #
# Date: February 2010 #
# Description: This SP-system models the sender cell strain developed by Ron Weiss group #
SPsystem senderCell
# Molecular species present in the system #
alphabet
PLtetO1_geneLuxI
proteinLuxI
proteinLuxI_Rib
rnaLuxI
rnaLuxI_RNAP
signal3OC6
endAlphabet
# Compartments of the system #
compartments
bacterium
endCompartments
# Initial number of molecules in the compartments #
initialMultisets
initialMultiset bacterium
PLtetO1_geneLuxI 1
endInitialMultiset
endInitialMultisets
# Molecular interactions produced by the synthetic circuit #
ruleSets
ruleSet bacterium
# Transcriptional fusion between the PLtetO1 and geneLuxI gene #
PLtetO1({LuxI},{0.1},{bacterium}) from promoterLibrary.plb
# Post-transcriptional processes associated with LuxI #
PostTransc({LuxI},{3.36,0.0667,0.004,3.78,0.0667},{bacterium}) from postTranscriptionalLibrary.plb
# Signal synthesis #
r1: [ proteinLuxI ]_bacterium -c1-> [ proteinLuxI + signal3OC12 ]_bacterium c1 = 5
# Signal diffusion #
Diffusion({3OC6},{2},{bacterium}) from postTranscriptionalLibrary.plb
endRuleSet
endRuleSets
endSPsystem
The bacterial strain, pulsingCell, producing a pulse of GFP protein as a response to the signal 3OC6-HSL (AHL) is modelled using the SP-system model below:
# Author: Francisco J. Romero-Campero #
# Date: February 2010 #
# Description: This SP-system models the pulsing cell strain developed by Ron Weiss' group #
SPsystem pulsingCell
# Molecular species present in the system #
alphabet
CI2
LuxR2
PluxL_geneLuxR
PluxPR_CI2_geneGFP
PluxPR_LuxR2_CI2_geneGFP
PluxPR_LuxR2_geneGFP
PluxPR_geneGFP
PluxR_LuxR2_geneCI
PluxR_geneCI
proteinCI
proteinCI_Rib
proteinGFP
proteinGFP_Rib
proteinLuxR
proteinLuxR_3OC6
proteinLuxR_Rib
rnaCI
rnaCI_RNAP
rnaGFP
rnaGFP_RNAP
rnaLuxR
rnaLuxR_RNAP
signal3OC6
endAlphabet
# Compartments in the system #
compartments
bacterium
endCompartments
# Initial number of molecules present in the system #
initialMultisets
initialMultiset bacterium
PluxL_geneLuxR 1
PluxR_geneCI 1
PluxPR_geneGFP 1
endInitialMultiset
endInitialMultisets
# Molecular interactions produced by the synthetic gene circuit #
ruleSets
ruleSet bacterium
# Transcriptional fusion between the PluxL and the LuxR gene #
PluxL({LuxR},{0.1},{bacterium}) from promoterLibrary.plb
# Post-transcriptional processes associated with LuxR #
PostTransc({LuxR},{3.2,0.3,0.04,3.6,0.075},{bacterium}) from postTranscriptionalLibrary.plb
# Dimerisation of LuxR in the presence of 3OC6 #
DimSig({LuxR,3OC6,LuxR2},{1,0.0154,1,0.0154},{bacterium}) from postTranscriptionalLibrary.plb
# Transcriptional fusion between the PluxR and the CI gene #
PluxR({CI},{1,1,4},{bacterium}) from promoterLibrary.plb
# Post-transcriptional processes associated with CI #
PostTransc({CI},{3.2,0.02,0.04,3.6,0.1},{bacterium}) from postTranscriptionalLibrary.plb
# Dimerisation of CI #
Dim({CI,CI2},{1,0.00554},{bacterium}) from postTranscriptionalLibrary.plb
# Transcriptional fusion between the PluxPR and the GFP gene #
PluxPR({GFP},{1,1,1,1,5,0.0000001,5,0.0000001,4},{bacterium}) from promoterLibrary.plb
# Post-transcriptional processes associated with GFP #
PostTransc({GFP},{3.36,0.667,0.04,3.78,0.0667},{bacterium}) from postTranscriptionalLibrary.plb
# Diffusion of signal 3OC6 #
Diffusion({3OC6},{0.1},{bacterium}) from postTranscriptionalLibrary.plb
endRuleSet
endRuleSets
endSPsystem
For technical reasons, our model using an extra cell to represent the boundary of the system:
SPsystem Boundary
alphabet
signal3OC6
endAlphabet
compartments
cell
endCompartments
initialMultisets
initialMultiset cell
endInitialMultiset
endInitialMultisets
ruleSets
ruleSet cell
r1: [ signal3OC6 ]_cell -c1-> [ ]_cell c1=1
endRuleSet
endRuleSets
endSPsystem
The geometry of a bacterial colony of the cell type or bacterial strain represented in the previous model is captured using the following rectangular lattice:
# Author: Francisco J. Romero-Campero #
# Date: February 2010 #
# Description: A rectangular lattice of size 11x31 #
lattice rectangularLattice
# Dimension of the lattice and lower/upper bounds #
dimension 2
xmin 0
xmax 10
ymin 0
ymax 30
# Parameters used in the definition of the rest of components defining the lattice #
parameters
parameter b1 value = 2
parameter b2 value = 1
endParameters
# Basis vector determining the points in the lattice #
# in this case we have a rectangular lattice #
basis
(b1,0)
(0,b2)
endBasis
# Vertices used to determine the shape of the outmost membrane #
# of the SP systems located on each point of the lattice #
vertices
(b1/2,b2/2)
(-b1/2,b2/2)
(-b1/2,-b2/2)
(b1/2,-b2/2)
endVertices
# Vectors pointing at the neighbours of each point of the lattice #
neighbours
(1,0) (1,1) (0,1) (-1,1)
(-1,0) (-1,-1) (0,-1) (1,-1)
endNeighbours
endLattice
Finally, the model of the synthetical bacterial colony is obtained by distributing cellular clones of the sender cell strain at one end of the lattice and cellular clones of the pulsing cell strain over the rest of the points. This is modelled using the LPP-system below:
# Author: Francisco J. Romero-Campero #
# Date: February 2010 #
# Description: This LPP-system models the spatial distribution of the different #
# cellular strains in the synthetic bacterial colony developed by #
# Ron Weiss' group exhibiting pulse propagation#
LPPsystem pulsePropagation
# List of the different bacterial strains used in the system #
SPsystems
SPsystem senderCell from senderCell.sps
SPsystem pulsingCell from pulsingCell.sps
SPsystem boundaryCell from boundaryCell.sps
endSPsystems
# The geometry of the system is determine using a regular finite point lattice #
lattice rectangular from rectangular.lat
# Spatial distribution of the cells over the lattice #
spatialDistribution
# The boundary of the system is filled with boundary cells #
positions for boundaryCell
parameters
parameter i = 0:1:30
parameter j = 0:10:10
endParameters
coordinates
x = i
y = j
endCoordinates
endPositions
positions for boundaryCell
parameters
parameter i = 0:30:30
parameter j = 1:1:9
endParameters
coordinates
x = i
y = j
endCoordinates
endPositions
# Sender cells are distributed at one end of the lattice #
positions for senderCell
parameters
parameter i = 1:1:5
parameter j = 1:1:9
endParameters
coordinates
x=i
y=j
endCoordinates
endPositions
# Pulsing cells are distributed over the rest of the lattice #
positions for pulsingCell
parameters
parameter i=6:1:29
parameter j=1:1:9
endParameters
coordinates
x=i
y=j
endCoordinates
endPositions
endSpatialDistribution
endLPPsystem
Simulations¶
Stochastic simulations of our model of the pulse generator can be run using the Infobiotics workbench. For this, please load using the provided interface the simulation parameter file, pulsePropagation.params, provided with the files comprising this example. Be patient, these simulations could take around five minutes.
Below we show a picture of the spatial propagation of a pulse of GFP over the bacterial colony. You can see our video here or here.
Model Checking¶
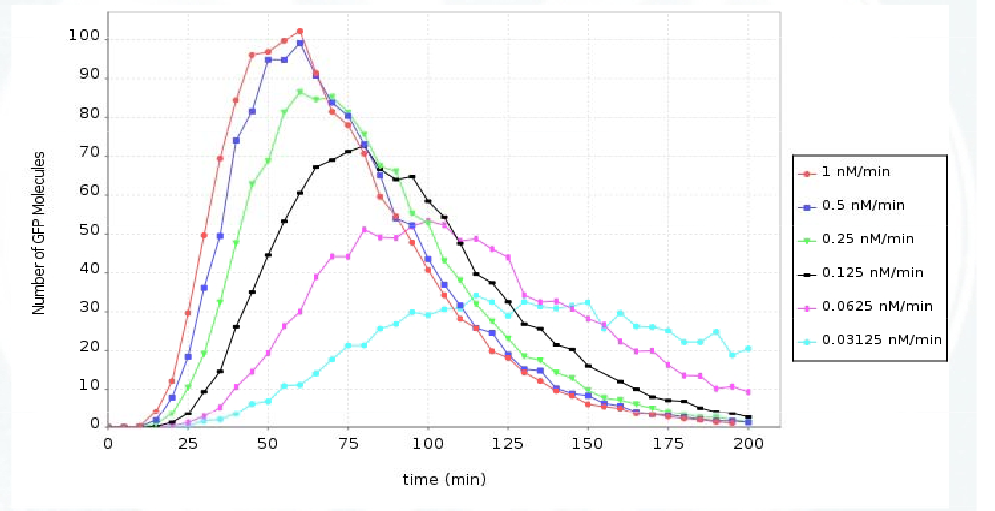
We observe that the further away the pulsing cells are from the sender cells the less likely they are of producing a pulse. We analyse this property by model checking the temporal formula below for a single pulsing cell recieving the signal at different rates:
R = ? [ I = T ] T = 0:5:200